fig3
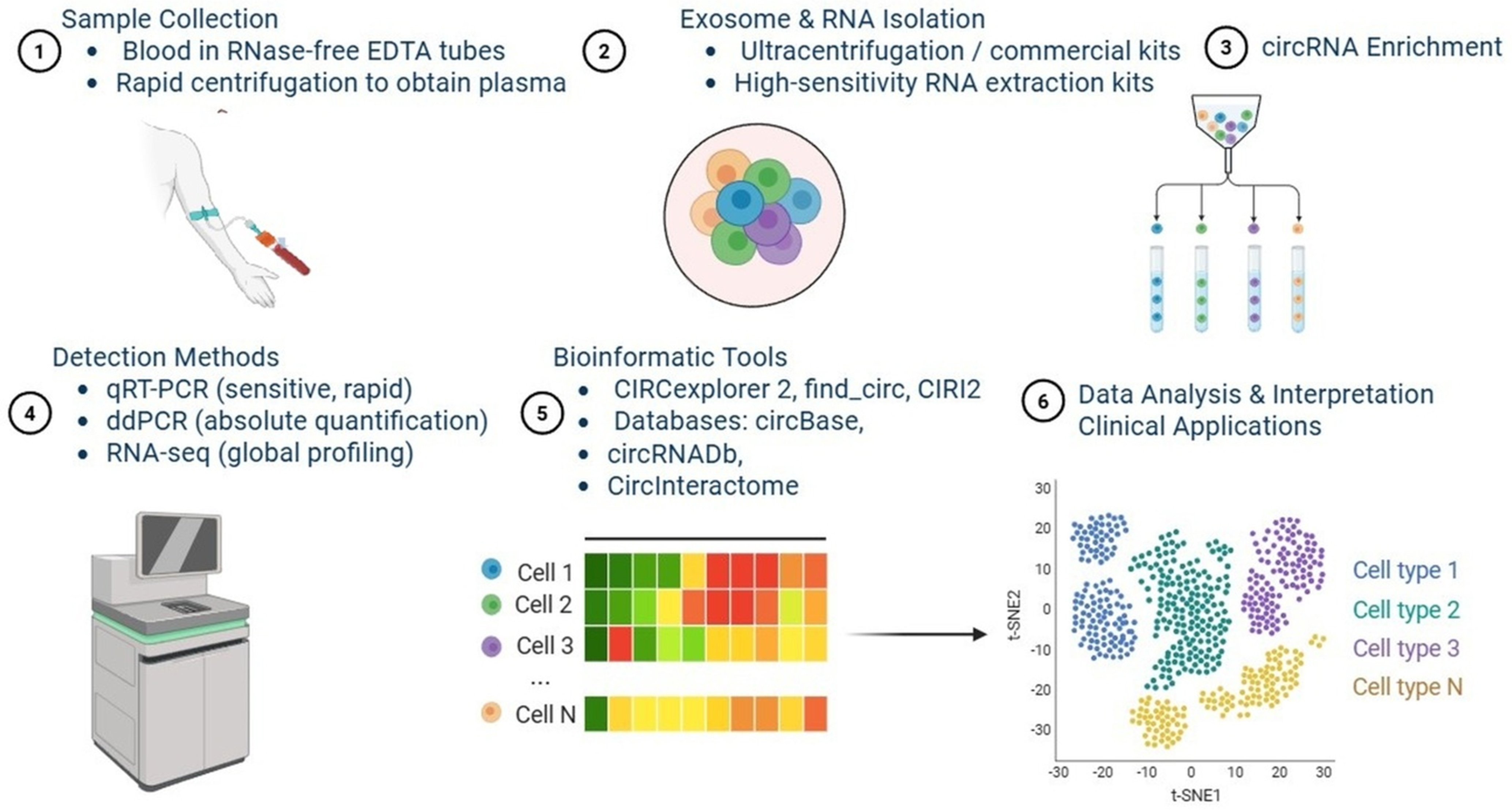
Figure 3. Schematic representation of the experimental and analytical pipeline for circRNA detection and characterization. (1) Sample collection: blood is collected in RNase-free EDTA tubes, followed by rapid centrifugation to obtain plasma; (2) Exosome and RNA isolation: performed using ultracentrifugation or commercial high-sensitivity RNA extraction kits; (3) CircRNA enrichment: strategies to enrich circRNAs for downstream analyses; (4) Detection methods: include qRT-PCR (sensitive, rapid), ddPCR (absolute quantification), and RNA-seq (global profiling); (5) Bioinformatic tools: circRNA analysis pipelines such as CIRCexplorer2, find_circ, and CIRI2, along with databases such as circBase, circRNADb, and CircInteractome; (6) Data analysis and interpretation: application of circRNA data in clinical translation and biomarker development [Created in BioRender. Singh DD (2025)]. circRNA: Circular RNA; EDTA: ethylenediaminetetraacetic acid; qRT-PCR: quantitative reverse transcription polymerase chain reaction; ddPCR: droplet digital polymerase chain reaction; RNA-seq: RNA sequencing.