fig2
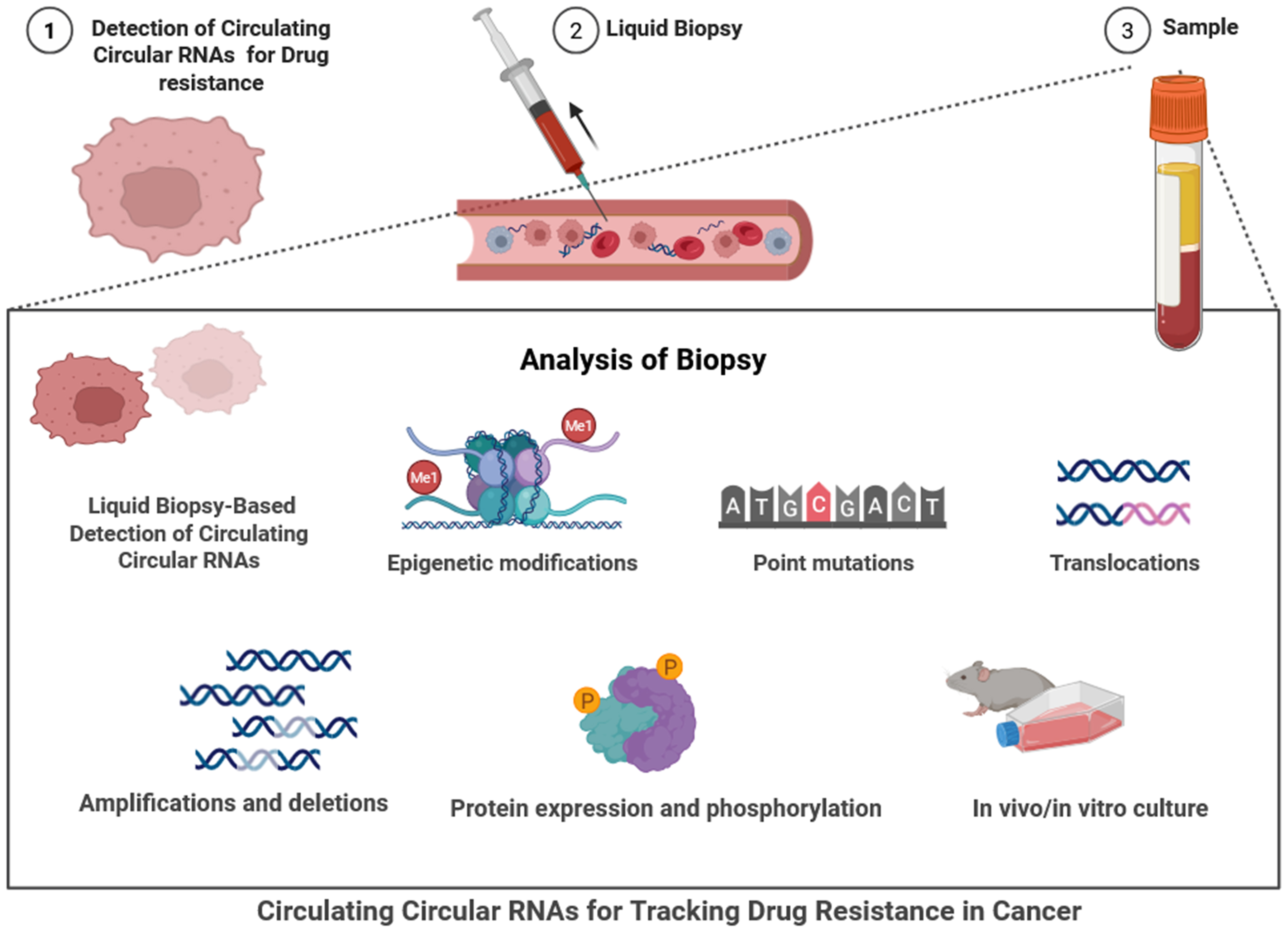
Figure 2. The figure depicts the use of liquid biopsy-based detection of circulating circRNAs to track drug resistance in cancer, beginning with the identification of drug-resistant tumor cells (Step 1) and progressing to liquid biopsy collection via blood draw and non-invasive material (Step 2). The resulting sample (Step 3), generally plasma or serum, will undergo analysis for molecular changes associated with the resistance. The analysis options include various molecular methods to detect epigenetic modifications, point mutations, translocations, and copy number changes such as amplifications or deletions. Other assessments also include protein expression and phosphorylated proteins, as well as the use of in vivo/in vitro methodologies to establish functional changes. Importantly, the aim is to detect and characterize the stable, circulating circRNAs in the liquid biopsy, where they may serve as potential circulating biomarkers. The proposed workflow enables real-time, non-invasive capture of valuable information regarding therapeutic resistance and supports the implementation of precision oncology and therapies for cancer treatment [Created in BioRender. Singh DD (2025)]. circRNAs: Circular RNAs.