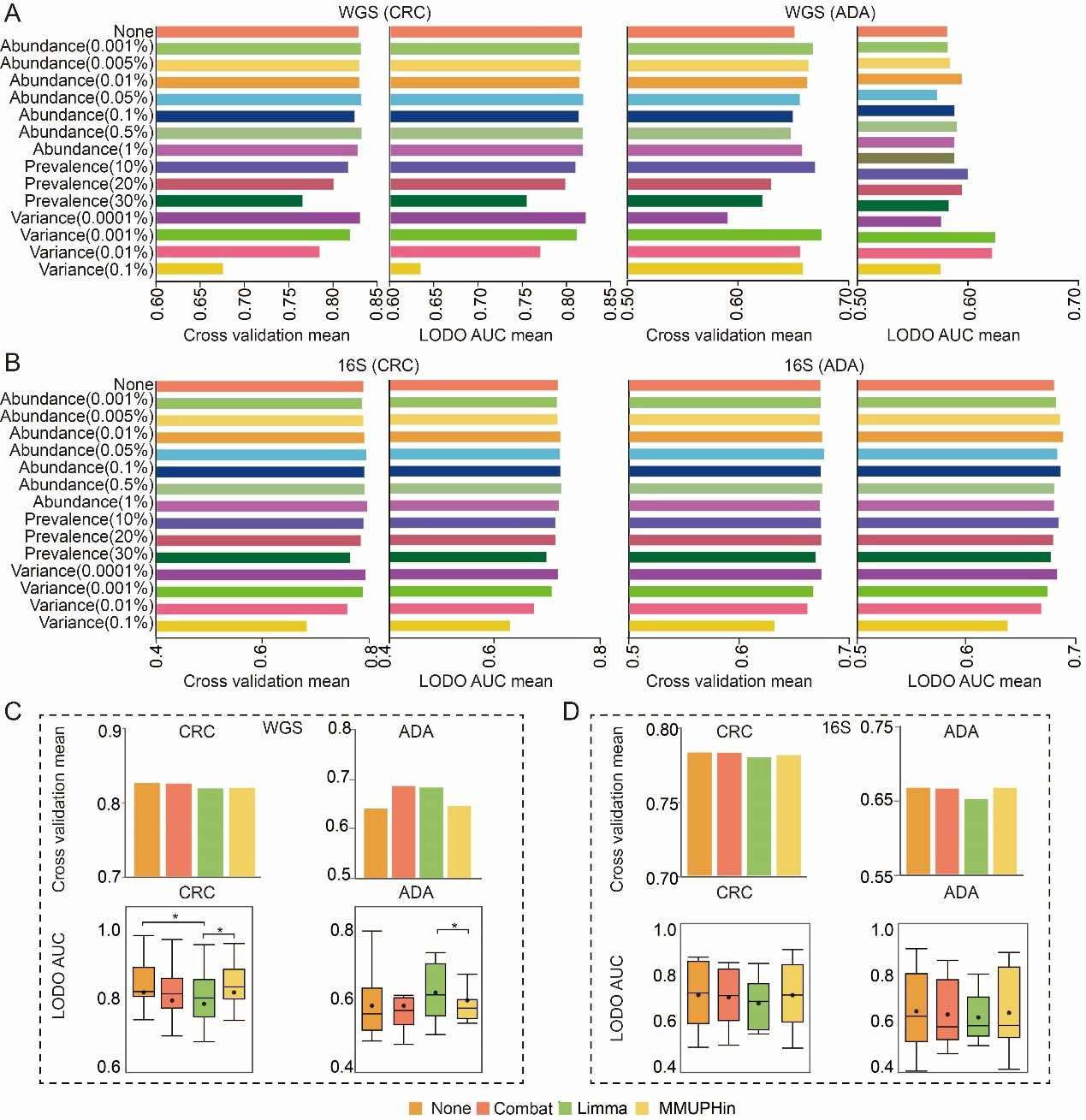
fig3
Figure 3. Exploration of optimal data preprocessing methods. (A and B) Comparison of 14 feature filtering thresholds against an unfiltered baseline on (A) WGS-SGB and (B) 16S-ASV data. Bars show the mean AUC from five-fold cross-validation (CV) and LODO validation; (C and D) Performance comparison between uncorrected models and three distinct batch correction methods on (C) WGS-SGB and (D) 16S-ASV data. Bar charts show the mean CV-AUC, and box plots illustrate the distribution of LODO-AUCs. Statistical significance against the baseline (uncorrected) was determined using a two-sided Wilcoxon rank-sum test (* P-value < 0.05). Source data is available in Supplementary dataset. WGS: whole genome sequencing; CRC: colorectal cancer; ADA: adenoma; 16S: 16S rRNA gene sequencing; AUC: area under the curve; LODO: Leave-One-Dataset-Out; Combat: ComBat (Empirical Bayes Batch Correction); Limma: linear models for microarray data; MMUPHin: meta-analysis methods with uniform pipeline for heterogeneity integration; SGB: species-level genome bin; ASV: amplicon sequence variant.