fig5
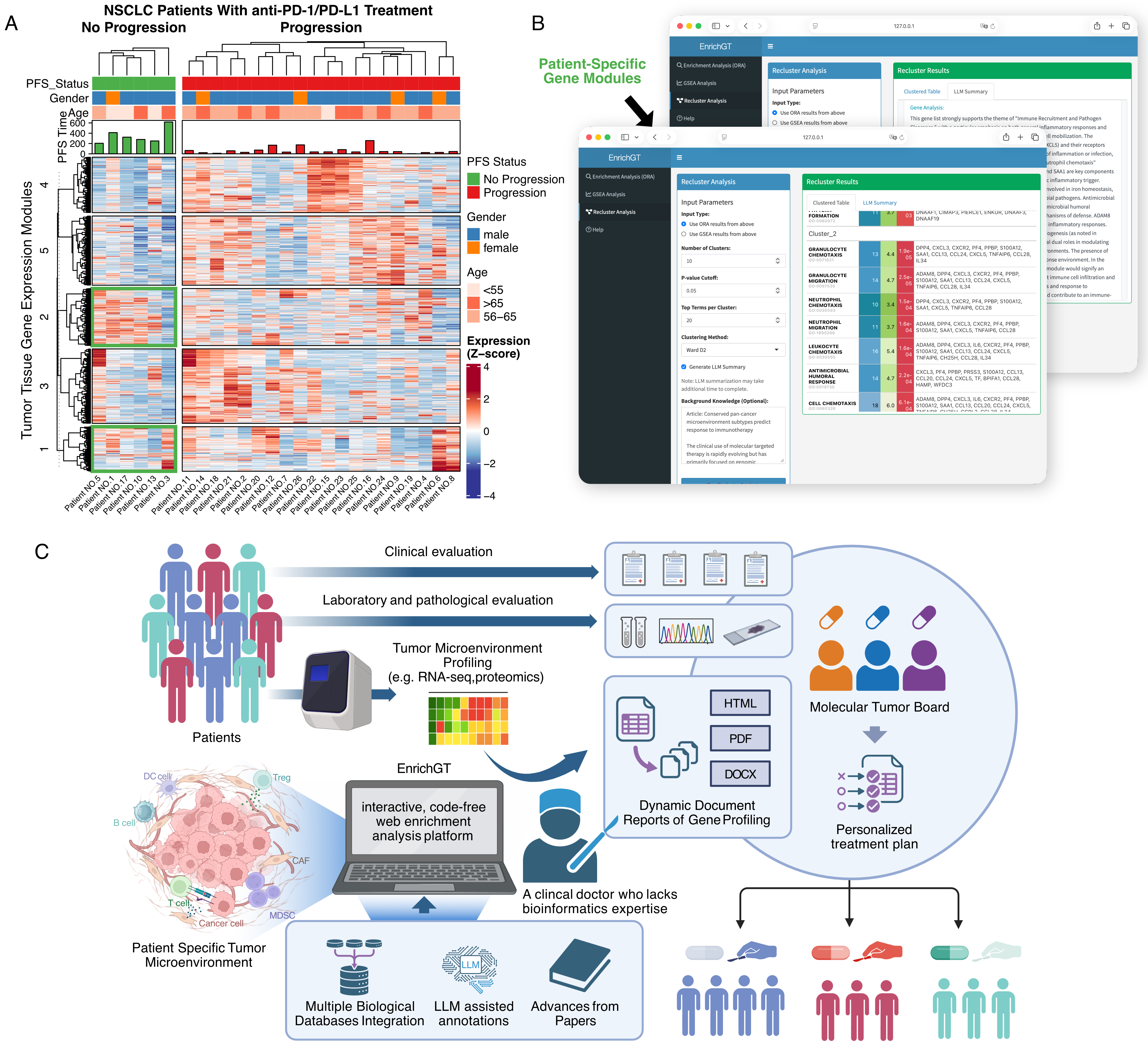
Figure 5. EnrichGT enables clinically interpretable, privacy-preserving analysis of immunotherapy response in NSCLC. (A) Heatmap of bulk RNA-seq profiles from tumor tissues of non-small cell lung cancer (NSCLC) patients treated with anti-PD-1/PD-L1. Columns represent individual patients grouped by progression-free survival (PFS; no progression vs. progression). Rows represent tumor tissue gene-expression modules identified by hierarchical clustering. Top annotation bars indicate PFS status, gender, and age group; bar plots show PFS time. Gene-expression values are displayed as Z-scores; (B) Example of the EnrichGT graphical interface running locally on a clinician’s computer. Patient-specific gene modules derived from bulk RNA-seq are uploaded and analyzed without the need for coding. The platform summarizes module-level enrichment results into interpretable tables and pathway maps, highlighting dysregulated pathways and tumor microenvironment features for each patient; (C) Schematic of the proposed clinical workflow. Patients undergo clinical, laboratory, and pathological evaluation followed by tumor microenvironment profiling (e.g., RNA-seq, proteomics). The resulting data are analyzed with EnrichGT, an interactive, code-free enrichment analysis platform that integrates multiple biological databases, LLM-assisted annotations, and up-to-date literature. EnrichGT generates dynamic gene-profiling reports (HTML/PDF/DOCX) that can be reviewed by molecular tumor boards and practicing clinicians, facilitating standardized interpretation of complex molecular data and supporting personalized treatment planning for individual patients. “Figure 5C” created in BioRender. xwk, g. (2026). https://BioRender.com/5o1azy5. PD-1: Programmed cell death protein 1; PD-L1: programmed cell death ligand 1; PFS: progression-free survival; LLM: large language model; DC cell: dendritic cell; B cell: B lymphocyte; T cell: T lymphocyte; CAF: cancer-associated fibroblast; MDSC: myeloid-derived suppressor cell; Treg: regulatory T cell; HTML: hypertext markup language; PDF: portable document format; DOCX: office open XML document; XML: extensible markup language; Z-score: standard score; ORA: over-representation analysis; GSEA: Gene Set Enrichment Analysis.