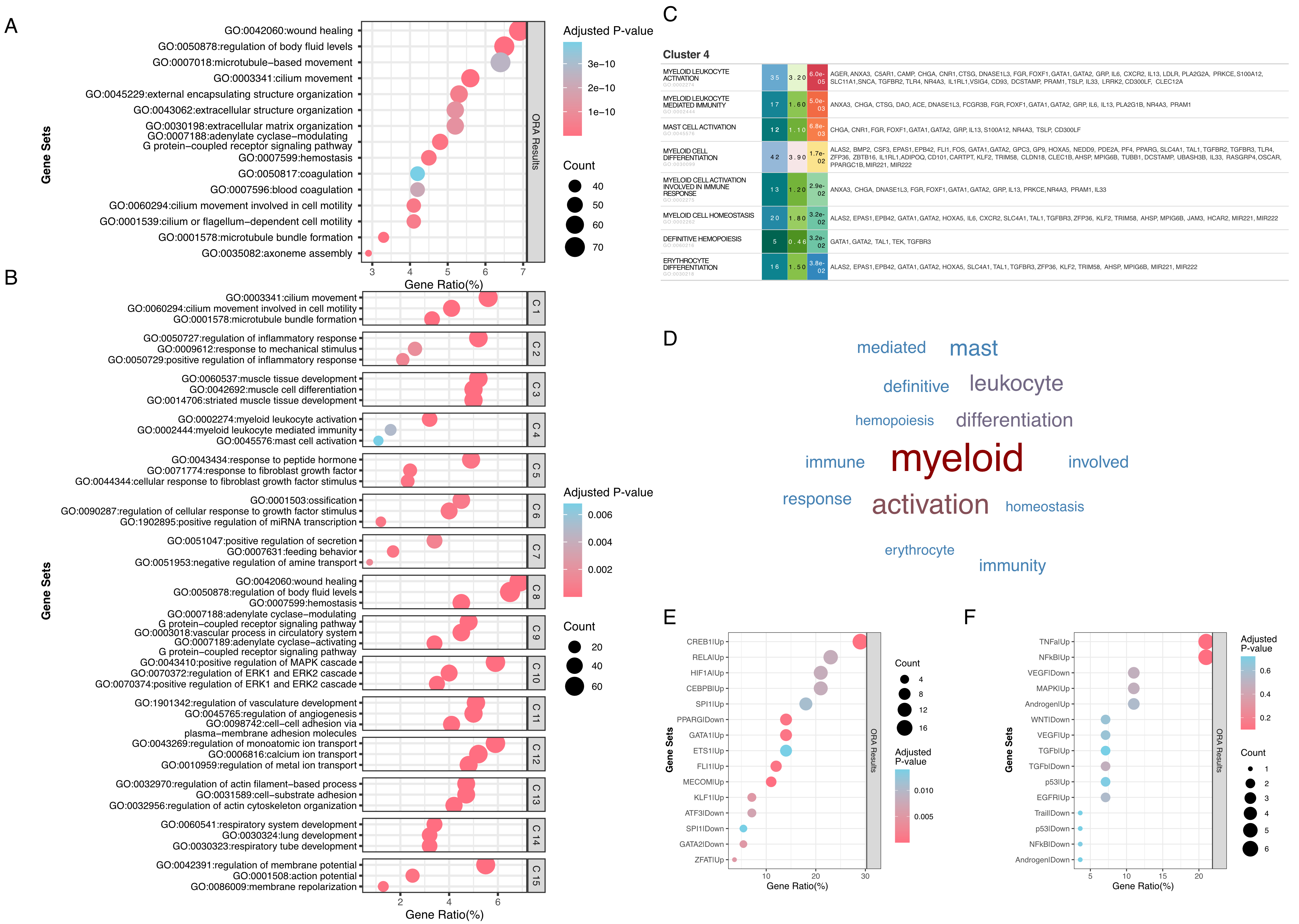
fig1
Figure 1. EnrichGT enables comprehensive biological interpretation through term clustering and dynamic visualization. (A) Traditional over-representation analysis of differentially expressed genes between high-stage (≥ Stage III) and Stage I lung adenocarcinoma from TCGA, showing the top 20 Gene Ontology Biological Process terms ranked by gene ratio. Bubble size represents the number of genes (count) enriched in each GO term; color intensity indicates the adjusted P-value. The x-axis shows gene ratio (percentage of genes in each term relative to the total query genes); (B) EnrichGT clustering of enriched terms into 15 biologically coherent clusters, with node size proportional to the number of terms per cluster and color intensity indicating enrichment significance (adjusted P-value); (C) Interactive table for Cluster 4 (myeloid cell activation), displaying all enriched terms with their enrichment statistics, enabling comprehensive data exploration within a single interface; (D) Word cloud visualization of Cluster 4 highlighting dominant biological themes, with word size proportional to term frequency; (E) Transcriptional regulator analysis of Cluster 4 genes using CollecTRI database, revealing key transcription factors driving myeloid activation signatures. Node size indicates the number of target genes; (F) Pathway activity inference for Cluster 4 using PROGENy, showing activation of inflammatory and hypoxia-related signaling pathways characteristic of tumor-associated macrophages (TAMs). TCGA: The Cancer Genome Atlas; GO: Gene Ontology; CollecTRI: Collection of Transcription Regulation Interactions; PROGENy: Pathway RespOnsive GENes for activity inference.